Studies and descriptors. - In essence, any biological study, be it experimental or observational, involves examination of groups, individuals, or parts of organisms, or of materials originally derived from organisms. Results of studies may take the form either of a series of values for defined parameters, or of unstructured textual information conveying a law or abstraction derived from the facts revealed during the investigation. Although unstructured textual information may also be stored, research databases lend themselves principally to the storage of information which may be expressed as parameter and value (characters and character states, "Descriptors"), because in this area the strength of electronic processing of large datasets takes effect. For the purpose of this article, comparative studies may be regarded as the processing of information gathered - and stored - as the result of individual studies.
From the point of view of the information modeller, the "Study" provides the framework to link descriptors with the organismic object of the study. Methods, persons, or bibliographical data related to the investigation are here registered. CDEFD used karyological investigations as an example to illustrate a complex type of study by means of a detailed information model (Berendsohn & al. 1996a). In contrast to the extensive descriptor structures described there, Study structures may take a rather simple form, e.g. in the recording of presence/absence data for floristic mapping. The function of the Study - entity may even be reduced to a link to a bibliographic reference. In any case, the Study represents the description and the result of an investigative process, which acts upon Biological Objects.
The "Biological Object". - A Biological Object is here defined as an entity-type or supertype in an information model. It provides a gateway between investigative or descriptive data and the organismic objects a defined study investigates. In a distributed database environment, the Biological Object may be used to provide a simplified view of the model to people who want to use a system based on it without knowing the intricacies of its design. Of course, in a relational system external information may be linked to many points, i.e., any of the entities may contain a key which can be used to link information in another, external entity to it. However, a defined interface has to be provided to people who either do not want to dive too deeply into the model's design, or who do not want to link their information too intimately with the collection system. For these, the Biological Object serves as a "switched" interface to link their information e.g. to the collection and taxonomic information covered by the IOPI and CDEFD models.
In the course of the investigation, the object of the study is initially always a material one: The animal which is observed, the soil sample containing microorganisms, the cell culture, or the tree in the forest under investigation. In the course of formulating results, the biological object may become an abstraction, e.g. a plant name representing a taxon, or an ecological category (a site investigated, a syntaxon, a biogeographical classification unit, etc.). Fig. 1 depicts the principal subdivision of Biological Objects.
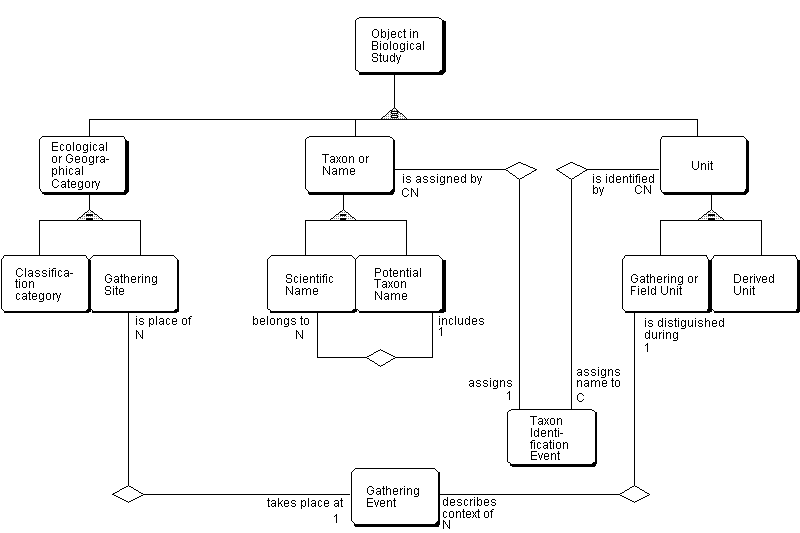
In the diagram, entity-types are represented by rectangles. They may be thought of as representing a table or a system of tables in a relational database. The triangles represent exclusive classification relationships (subtyping). An Object in a Biological Study (supertype) may either of the following subtypes: an Ecological or Geographic Category, a Taxon or Name, or a Unit. Other relationships are read along the connecting lines, starting with the entity-type name, followed by the descriptive text nearest to it, then the cardinality (i.e., how many instances of the second entity-type are referred to the instance in the first one) and, finally, the name of the second entity-type. The cardinality may be "1" (exactly 1), "C" (0 or 1), "N" (1 to many), or "CN" (0 to many). The "C" is for conditional relationship, i.e. it is possible that no instance is referred to. For further details on modelling techniques please refer to Berendsohn & al. (1996b). Ecological categories. - Because of the great diversity of investigative approaches and classification systems involved, it may prove impossible to provide a generalized information model for ecological categories. However, as in comparative studies, the basic data needed for such an investigation may well take the form of individual studies on organisms, related to one another, or related to a specific position in space and time (a site). This area is in urgent need of more basic research.
Names and taxa. - Using taxon names as biological objects in a study may be problematic, because a name may represent several concepts of a taxon. At least two cases must be distinguished: the name is provided on its own, or in the form of a "Potential Taxon" (Berendsohn 1995), i.e. bibliographic or other references are provided which clarify the taxonomic concept represented by the name. Much thought has been given to the structure of taxon name information (e.g. Beach & al. 1993), standards (Bisby 1994) and detailed information models have been developed (e.g. Berendsohn 1994).
Units. - CDEFD's main concern were material biological objects, which are referred to as Units. Fig. 1 illustrates the principal relationships of units in the CDEFD model, details can be found in Berendsohn & al. (1996b). Two main categories (subtypes) of Units are recognized:
The "Gathering or Field Unit" represents the biological object in its original location, unaltered by the investigative process. The entity-type "Gathering Event" provides information concerning the Who and When of the observation of the object, and it links units to the "Gathering Site", which in turn (directly or indirectly) provides all relevant locality data.
The second subtype is the "Derived Unit". Units may be derived from other Units, both, field and derived units. E.g. a microscopic slide may be prepared from a fungus found on a leaf which has been taken from a herbarium sheet. Each of these items (herbarium sheet, leaf with fungus spores, microscopic slide) do form a derived unit which may have distinct information attached to it (e.g. the taxonomic determination differs for fungus and herbarium sheet; the storage location may be different for all three items, etc.). By means of the gathering site stored with the original unit (the tree from which the herbarium sample was taken), the original location can be named for all these items. A Derived Unit is the product of a "Derived Unit Creation Event", which may be a process of curation, preparation, cultivation, or a transfer event, which creates one or more Derived Units from one (or rarely more) parent unit(s). As the model allows multiple iterations of this operation, it permits to store highly iterative processes, such as cultivation and propagation histories.