 |
Biodiversitätsinformatik / Biodiversity Informatics |
 |
Biodiversitätsinformatik / Biodiversity Informatics |
PREPRINT, to be published in: G. Curry & C. Humphries (ed.): Biodiversity Databases. Download as PDF.
Networking taxonomic concepts – uniting without "unitary-ism"
Walter G. Berendsohn & Marc Geoffroy
Botanic Garden and Botanical Museum Berlin-Dahlem, Freie Universität Berlin, Königin-Luise-Str. 6-8, 14195 Berlin, Germany
Introduction
One of the principal aims of current efforts in biodiversity informatics is to network the electronically available information about organisms from a wide variety of sources. This information has been produced at different times and places and with differing aims and is normally pigeonholed by means of the organism’s scientific name. However, correct (accepted) names are formed according to rules of nomenclature, without regard to the concept or circumscription of the taxon itself (Berendsohn 1995). Potentially, correct names stand for differing concepts (potential taxa). Consequently, names are not providing a reliable index for biodiversity information – but electronic networks such as the Global Biodiversity Information Facility or the European BioCASE (see Scoble, this volume) do need such an index for information access.
That taxonomic concepts pose a problem for taxonomic databases has been recognized already a decade ago (e. g. Beach & al. 1993). In contrast to information provision mediated by people (normally specialists), public databases need integrated explicit knowledge to reliably transmit complex information. In the 1990ies, information models laid the theoretical base for handling taxonomic concepts – names used in the sense of a certain circumscription (Zhong & al. 1996, Berendsohn 1997, Pullan & al. 2000, Le Renard 2000, Ytow & al. 2001, Anonymous 2003). Later, Software developed also demonstrated the practicability of such models for taxonomic data (e. g. Pullan & al. 2000, Gradstein 2001). This article summarises and updates results from our working group published earlier (Berendsohn 2003), states further evidence for the relevance of the problem, and reports progress made with the implementation of the Berlin-Model (Berendsohn & al. 2003).
Many still doubt that there is a need for the representation of concepts in taxonomic databases. For the technical implementation, it is certainly easier to do without. Even taxonomists often tend to think only of the basic science side of their endeavour, i. e. the product of their work - the latest taxonomic treatment - should be regarded as the state of the art and earlier works should stand corrected. However, this disregards the fact that names of organisms are widely used by non-taxonomists and have been used so for a considerable amount of time. So it is not a hobby of the information modeller’s community, but a duty of the systematics community to ask the questions how stable concepts are in taxonomy and how reliable names are as an index to biodiversity information.
The answer is that for many groups we don’t have a clue, in some groups we know that there is considerable stability (especially if supported by the nomenclatural methodology, e.g. in bacteriology), and in others we suspect that there is a high degree of instability, but we lack hard data. Explicit statements of concept conflicts (and lack thereof!) are rare and mostly hidden in monographer’s notes etc. Two recent publications, both for groups of plants, provide data to assess concept stability and thus the extent of the problem.
Assessing concept stability
For an analysis of concept stability, we need more data than contained in traditional checklists with their lists of synonyms (although these can also be used for analysis, see Geoffroy & Berendsohn 2003). We need datasets where concept relationships have been recorded in a comprehensive way, i. e. for every taxon treated, rather than in the largely anecdotal way found in traditional treatments. In analysing such datasets we have to be conscious of the fact that the resulting values for stability strongly depend on three factors: (i) the selection of other works with which the current concept is compared, (ii) the quality of the comparison itself, and (iii) the degree of taxonomic creativity of the authors of the present work itself. While neither selection nor quality can be assessed in a quantitative way, the third factor, here called "novelty" can. We simply calculate the amount or percentage of concepts for which no congruent concept is cited in the compared existing literature. A low novelty value indicates a conservative treatment, which will yield values that depend less on the current treatment and more on the variability of concepts already in use.
The first example publication is the "Standard list of the ferns and flowering plants of Germany" (Wisskirchen & Haeupler 1998), which was and is maintained as a database at the German Federal Agency for Nature Protection (BfN 2004) and available on the World Wide Web via the Agency’s Flora Web portal. In the publication, 4709 accepted taxa are listed, 3811 of these are species, the rest infraspecific taxa. The novel feature of this work is that Wisskirchen and Haeupler indicate the relationship of their taxon concept with that in a number of contemporary floristic works commonly used by students and practitioners of the German flora to determine plant species (or properties of plant species). Figure 1 gives an excerpt to illustrate the data.
This work was clearly not carried out with the aim to analyse concept stability, and it only states congruence or non-congruence of concepts without finer details. Nevertheless, the fact that it is fully databased gave us the opportunity to attempt an analysis of the dataset. To start with the assessment of novelty, we think the numbers suggest that a conservative approach was taken: 62% of the taxon concepts in the list are also treated in all other works, for 93% at least one congruent concept is cited among the other works included in the comparison. As to concept and nomenclatural stability, the dataset confirms that roughly half of German vascular plant taxa are stable as to name and concept, and 60% as to concept, throughout the works compared.

Figure 1: An extract from Wisskirchen & Haeupler (1998, reproduced with kind permission by Verlag Eugen Ulmer). The main body of text is a typical botanical checklist, with the correct (accepted) names in boldface, an abbreviated citation of the publication of the name, and of the type specimen. The common name is followed by a list of synonyms, i. e. names that either have the same type, or names the type of which is considered to by included in the concept the correct name stands for.
The unusual feature of the work is embedded in the letter codes on the right, which stand for floristic works in current use in Germany. Uppercase letters indicate that the name is considered to be used with the same circumscription in those works, lowercase letters indicate a different concept. Missing letters indicate that the name was not used. For Anagallis arvensis, 6 works use the same name with same concept, 1 has a different concept. For Anagallis foemina 6 works use same name with same concept, 1 doesn’t use name nor the concept.
These results compare well to the data found for German mosses in a study much more focused on the concept issue. For their "Reference List of the Mosses of Germany", Koperski, Sauer, Braun & Gradstein (2000) used the concept-oriented IOPI model (Berendsohn 1997) for their data-recording. They related the 1548 accepted taxa to concepts in 11 floristic or taxonomic treatments, which they considered to be in current use (Fig. 2). Koperski & al. based their assessment on detailed comparison of descriptions and discussion of the concepts. The relationships between potential taxa (PT) documented represent the 5 basic relationships between two concepts that can be stated when the concept or potential taxon is perceived as a set of objects (specimens, observations, etc.): (i) PT1 and PT2 are congruent; (ii) PT1 is included in PT2; (iii) PT1 includes PT2; (iv) PT1 and PT2 overlap each other; and (v) PT1 and PT2 exclude each other.

Figure 2: An extract from Koperski & al. (2000, reproduced with kind permission by the German Federal Agency for Nature Protection). The top three lines represent a "traditional" checklist entry, with the correct name, its protologue citation (with an indication of the source), and a list of synonyms (here only one, the equal sign indicates that it is a heterotypic synonym). The following lines cite several potential taxa, i. e. names accepted in a certain reference (the citation following the "sec." for secundum, according to). The symbol in the beginning shows the concept relationship with respect to the accepted (correct) name: congruent, included in, including, and overlapping.
Of the 1.548 accepted potential taxa (PTs) in Koperski & al., 1.509 (97%) have one or more congruent concept(s) within the compared works. A low degree of novelty can be recognized.
With respect to concept stability, for 515 (33%) of the taxa listed, wider concept(s) have been found among the other treatments, for 267 (17%) included concepts, and for 90 (6%) overlapping concepts were identified. Including the facts that can be drawn from the traditional synonymic list, we come to the following result of the analysis of the moss dataset: With respect to nomenclatural and concept stability, 55% of the taxa show stability with respect to concepts, i.e. they cite no relations to other concepts but congruent ones. These 55% can be further subdivided: for 35% we can state that only homotypic synonyms have been cited, so that there remains little doubt about their stability. However, only 17% of all taxa listed offer that level of stability under a constant name. For 20% there is some indication of instability, e.g. there are heterotypic synonyms or misapplied names cited. As opposed to that, 45% of the taxa show explicit instability, i.e. there are other concept relationships cited than congruent ones (Fig. 3).
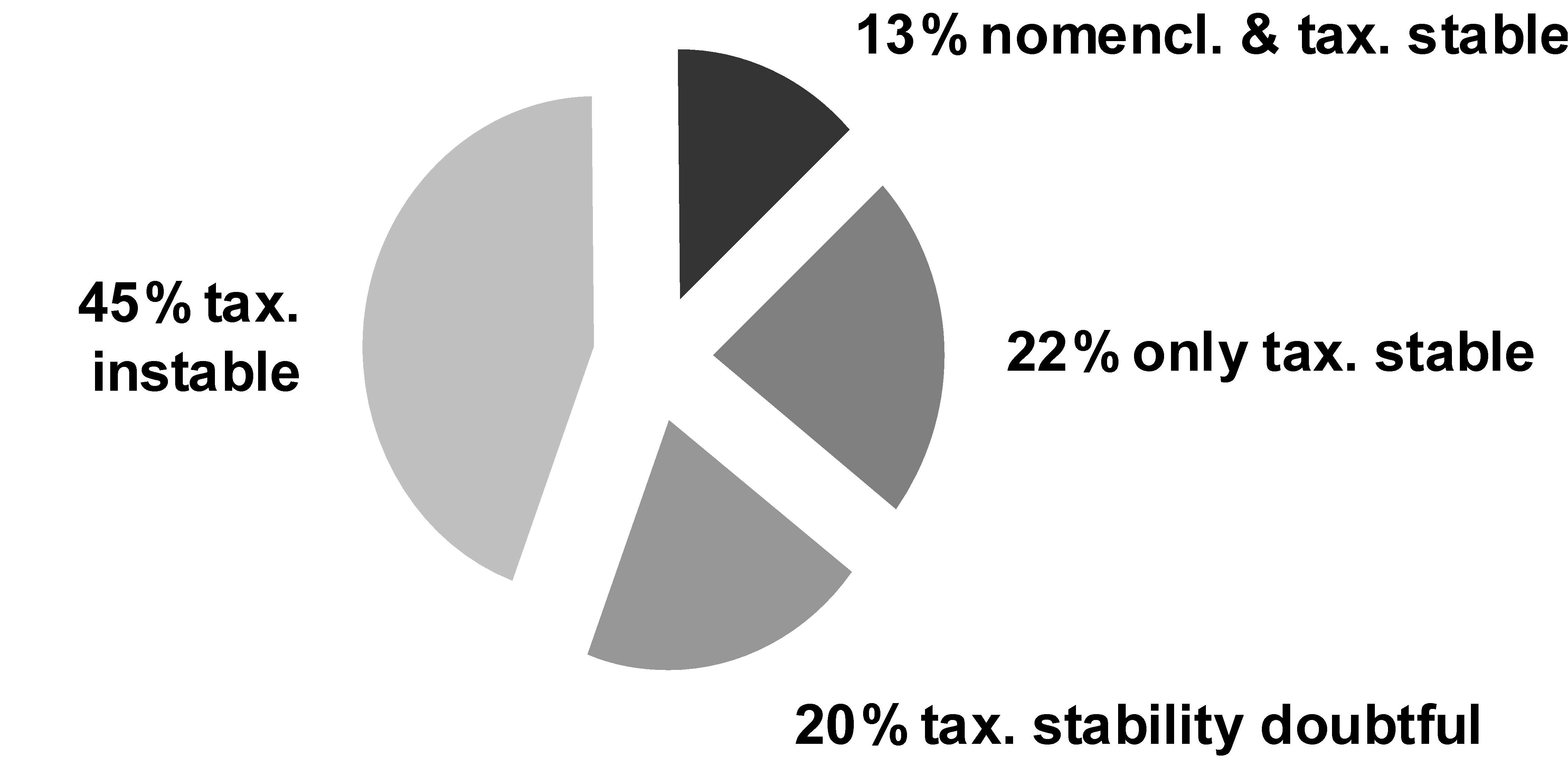
Figure 3: Nomenclatural and taxonomic stability in German mosses. Data from Koperski & al. 2000.
Summarizing, a considerable portion of plant taxa exist for which a high degree of instability of concepts even among works in current use can be affirmed. Content linked to taxon names includes, inter alia, uses (mostly human) and threats (to species itself, to hosts, to health, to environment, etc.), ecology (pollination, symbiosis, parasitism, indicator value, edaphic and climatic requirements, etc.) of species, molecular data (natural substances, genes, sequences, physiology, etc.), geographical range or occurrence, and descriptive data. Kirschner & Kaplan (2002) have strikingly demonstrated how compilation of lists of names can lead to inaccurate information of high practical importance (in that case, Red Lists of threatened plants). Considering the increasing ease with which these data can be linked using the Internet, and considering the obvious hazards of uncritical linking of such knowledge by means of taxon names, systematists have to take action to construct systems that more reliably inform users about the caveats (or lack thereof) of information linkage.
A concept-oriented system – current state
Fig. 4 depicts a system that relays information from providers of taxon-linked factual information to users querying on taxon names. This simple model can work given one of the following scenarios: all data providers agree on common concepts for the taxa involved, or each provider is treating a single taxonomic group only, for which they provide the authoritative view. The second scenario is followed by the Species 2000 system (Roskov & Bisby 2004), the first one is supposedly followed by the ITIS system in the US (ITIS 2004). We posit that, given the inherent problems in taxon concepts and naming, this system is suboptimal at least when legacy data, divergent views and general resource discovery is to be supported. The broker module must be supported by a series of additional modules that effectively mediate the information access via names and allow a dependable transmission of factual information.
The central component of such a system is a database (which may be distributed over several sites) that allows to store taxon concepts and their relationships. Such a system will allow the linkage of different information sources independent of the concept, and the transmission of information along the concept relationships established in the database. It will also allow calculating the dependability of a name as the representation of a concept, and thus allow making a conjecture on the concept stability for a name introduced without explicit concept relation (as most of the factual databases presently go).
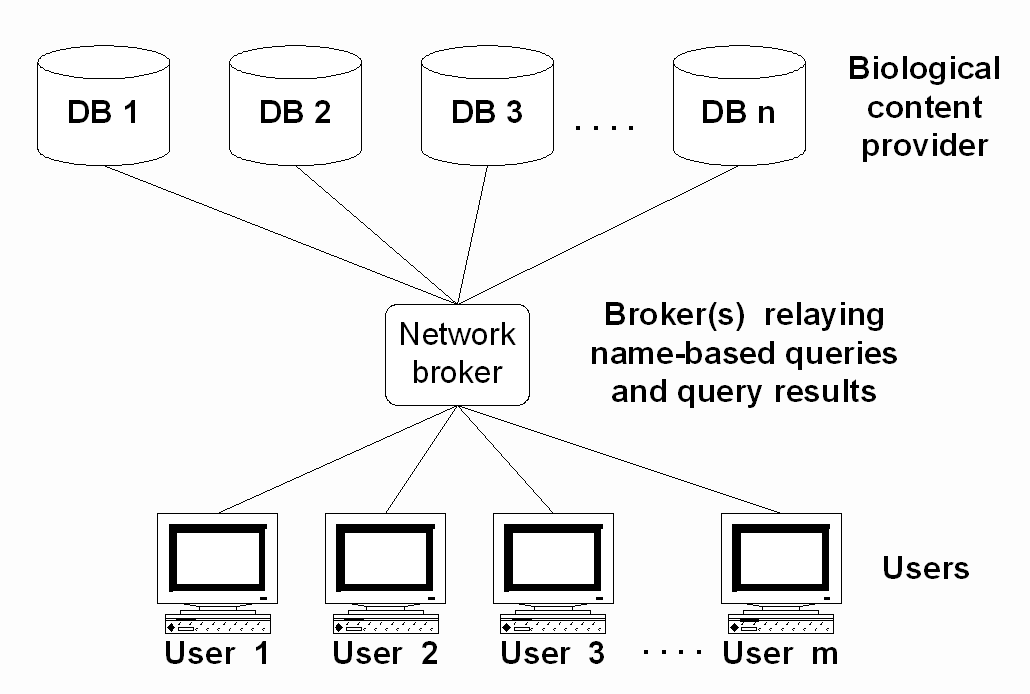
Figure 4: A simple model for access to information linked to taxon names.
Of course the content of this database has to be edited and kept up to date, so an editor software component is necessary. Preferably there should be one for local data maintenance, drawing on fast connections and the functionalities available in a local client-server environment, and a remote editor, to be able to edit the database over the Internet.
Finally, this database also needs output tools, both for print media and for the World Wide Web, supporting the tasks the editor of a taxonomic work would need, and so encouraging taxonomists to actually use and improve upon the data in the database.
With the "Berlin Model" database (Berendsohn & al. 2003) and related tools the database, editor and output tools have been put in place. The development has been supported by several projects, which share the core database model and functionality:
The EU funded Euro+Med PlantBase project supported the development of the Internet Editor software (Güntsch 2003). Another EU project, Species 2000europa currently supports the installation of Berlin model database and tools at the Euro+Med central site in Reading. The same project will also create a standardized access to both, the Euro+Med database in Reading and the IOPI database hosted in Berlin, which is also based on the Berlin Model. The IOPI database provides access to legacy datasets as well as checklist access to the taxonomic treatments published in the Species Plantarum series. For example, the treatment of Juncaceae (Kirschner 2002) is published there in its original form; at the same time, a parallel dataset is further improved and added to by the Juncaceae working group using the Remote Editor. Med-Checklist (vol. 2, Compositae) and the Dendroflora of El Salvador are two Berlin-based checklist projects currently using the database. The AlgaTerra project has been instrumental in developing the local editor software and devising a comprehensive extension of the system to cover type specimens and their assertion. AlgaTerra cooperates with 7 partners in German to link information from molecular investigations, herbarium specimens, and cultured strains via a common taxonomic core on microalgae (AlgaTerra 2004). The same approach is also followed by the German Federal Agency for Nature Protection. As mentioned before, the German standard list is available on-line (BfN 2004), however, up to now only as a single-concept checklist. Not without reason is it this user of taxonomic information pushing the development of concept-based checklists in Germany. The Agency primarily deals with information linked to names as opposed to the taxonomy itself. The standard list database is currently being converted to a Berlin Model database by the MoReTax project. This system will also be used as the taxonomic access system for information on German specimens and observations within the German GBIF-Node for Botany (GBIF-D 2004).
In technical terms, the databases are currently implemented under MS SQL-Server (and Oracle), with cross-project co-ordination of database-level functions and procedures. The Remote Taxonomic Editor was implemented using the ColdFusion application server and Java. The local taxonomic and extensions editor is based on Visual Basic, while database maintenance tools (data integrity checking mechanisms etc.) as well as the WWW output use various clients and tools.
A database as the taxonomic core, editor software to input and change data, and database maintenance tools are available and already allow us to produce and publish traditional as well as concept oriented checklists. We are now starting to meet the challenge to create a broker system incorporating the concept relationships present in a Berlin-Model database which acts as the system’s "taxonomic core".
The user may issue a query to get information about a certain taxon (name) from different sources (e.g. distribution information from one database, medical uses from another, and red list status from a third). Equally, the user may directly query the content (red-listed organisms with medical properties from Germany). In both cases, taxon names are used to produce the result, the second case only differs in that the names to be searched for are coming from the content databases themselves.
These databases may themselves specify a taxon concept as their taxonomic reference point, or only a taxon name. In the former case, matters are greatly simplified, because the content can be directly linked to a concept in the taxonomic core. The following account of the broker’s function will be based on the latter, currently prevailing, case.
The broker performs the following functions:
(i) it searches the taxonomic core database to retrieve all known names for the taxon
(ii) it gets the requested content linked to these names from the connected databases, and
(iii) it provides the content to the user, including statements to explain the way it has expanded the query in step (i) as well as caveats resulting from the taxonomic core’s knowledge about concept instabilities for that particular name. This presentation of content strongly depends on the level of expertise of the user, which should be defined to at least distinguish taxonomists from the rest of the world.
The broker should provide as much trustworthy information as possible to the user. This may be simple – in the case where the taxonomic core provides reasonable proof that all used names stand only for a single concept (all concepts are congruent; all synonyms are "unequivocal**" in Species2000 terminology). However, as we have shown above, this is not always the case even for a single specified name. Moreover, in many cases we still lack explicit statements as to concept relationships, and we have to rely on implicit information, such as that given in the taxonomic hierarchy (a subspecies "is included in" its species) or lists of synonyms (homotypic synonyms at least share their type, so their relationship is at least "overlapping").
The broker has to rely on a "transmission engine", a component responsible for selecting those concepts and names that are related to the given name and to which content information may have been linked. To disclose these relationships even where they have not been explicitly stated rules must be established that define on the one hand how far the engine should go in its processing, i. e. how "deep" the chain of possible consecutive relationships (from PT1 to PT2, from PT2 to PT3, …, PTn-1 to PTn) should be. On the other hand, rules define the relationship arising between PT1 and PTn according to the particular relationships involved in the chain between them and the nature of the information to be transmitted (see below). Further rules of the transmission engine specify which information and with which "caveats" should be displayed to the user depending on
(a) the resulting relationships to the concept to which this content was linked to
(b) the level of expertise of the user who issued the query and
(c) the "nature" of the information to be transmitted. For example, some information relates to every element in a taxon ("is a tree"), some to some elements ("has wings" in a taxon where larvae are wingless), and some to the entire set but not to individual elements ("occurring in Germany and France). Such classes of information require different processing in the transmission engine and different display.
In conclusion, to provide meaningful output the system must consider a complex set of rules and parameters for the construction and use of relationships between taxonomic concepts. It also needs to know about the nature of the information transmitted, and of the level of expertise of the user of the system. This information has to be stored as part of the broker’s transmission engine component, and an editing tool (the "rule tuner") must be implemented to be able to tweak the output of the system.
The theoretical base for these components was detailed by Geoffroy and Berendsohn in several articles in Berendsohn 2003. Presently we are starting to meet the challenge to create such a "transmission engine" and the "rule tuner". The combination of the concept-based taxonomic core database ("Berlin Model") and the "transmission engine" will help us to network and better utilize the growing number of available content providers for biodiversity information. First attempts to implement user interfaces with some of the "transmission engine" and "rule tuner" features are currently under way in the MoReTax and GBIF-D Botany projects, using the German Plant datasets, and in the AlgaTerra project.
Conclusions
Users demand a web-based "Unitary Taxonomy" (Godfray 2002) to get reliable access to species information. However, THE taxonomic revision is as a rule not possible, because local treatments, lack of new treatments, or different hypothesis’ lead to co-existing taxonomies (Scoble 2004). Using modern IT tools, taxonomists can easily provide information on concept relationships between different systems and treatments thus creating a pathway between current and past treatments. At the very least, specialists should make an effort to state where there appears to be no problem. Transmission models will allow using concept relationships, also those extracted from "traditional" synonyms and (perhaps) specimens, for an access system that relates information from different sources to the user. A concept-based taxonomic information system thus unites the taxonomic research process with reliable name-based user access to biodiversity information
Acknowledgements
The team in Berlin working on the software components mentioned also includes Andrea Hahn, Anton Güntsch, Javier de la Torre, Jinling Li, Karl Glück, Markus Döring, and Wolf-Henning Kusber
References
AlgaTerra 2004. AlgaTerra - An information system for terrestrial algal biodiversity: a synthesis of taxonomic, molecular and ecological information. [http://www.algaterra.net]
Anonymous 2003. VegBank Taxonomic Data Models. Ecological Society of America. [http://vegbank.org/vegbank/design/planttaxaoverview.html]
Beach, J. H., Pramanik, S. & Beaman, J. H. 1993. Hierarchic taxonomic databases. Ch. 15 (pp. 241-256) in: Fortuner, R. (ed.): Advances in computer methods for systematic biology: artificial intelligence, databases, computer vision. John Hopkings University Press, Baltimore.
Berendsohn, W. G. 1995. The concept of "potential taxa" in databases. Taxon 44: 207-212.
Berendsohn, W. G. 1997. A taxonomic information model for botanical databases: the IOPI model. Taxon 46: 283-309.
Berendsohn, W. G. 2003. MoReTax – Handling factual information linked to taxonomic concepts in biology. Schriftenreihe Vegetationsk. 39.
Berendsohn, W. G., Döring, M., Geoffroy, M., Glück, K., Güntsch, A., Hahn, A., Kusber, W.-H., Li, J.-L., Röpert, D. & Specht, F. 2003. The Berlin Taxonomic Information Model. Schriftenreihe Vegetationsk. 39: 15-42.
BfN 2004 [Mar 11]. Bundesamt für Naturschutz [Federal Agency for Nature Protection]. Floraweb – Daten und Informationen zu Wildpflanzen und zur Vegetation von Deutschland. [www.floraweb.de]
GBIF-D 2004 [Mar 11]. The GBIF Programme in Germany. [http://www.gbif.de/homeenglish]
Geoffroy, M. & Berendsohn, W. G. 2003. The concept problem in taxonomy: importance, components, approaches. Schriftenreihe Vegetationsk. 39: 5-13.
Gradstein, S. R., Sauer, M., Braun, M., Koperski, M., & Ludwig, G. 2001. TaxLink, a program for computer-assisted documentation of different circumscriptions of biological taxa. Taxon 50: 1075-1084.
Godfray, H.C. J. 2002. Challenges for taxonomy. Nature 417: 18-19.
Güntsch, A., Geoffroy, M., Döring, M., Glück, K., Li, J.-L., Röpert, D., Specht, F. & Berendsohn, W. G. (2003): The taxonomic editor. Schriftenreihe Vegetationsk. 39: 43-56.
ITIS 2004 [Mar 4]. Integrated Taxonomic Information System on-line database. About ITIS – background information. http://www.itis.usda.gov/info.html.
Kirschner, J. 2002 (ed.). Juncaceae 1-3. Species Plantarum: Flora of the World Part 6-8. Canberra.
Kirschner, J. & Kaplan, ˇ. 2002. Taxonomic monographs in relation to global Red Lists. Taxon 51: 155-158.
Koperski, M., Sauer, M., Braun, W. & Gradstein, S. R. 2000. Referenzliste der Moose Deutschlands. Schriftenreihe Vegetationsk. 34: 1-519.
Le Renard, J. 2000. TAXIS, a taxonomic information system for managing large biological collections. P. 18 in: Abstracts. TDWG 2000: Digitizing Biological Collections. Taxonomic Databases Working Group, 16th Annual Meeting, Frankfurt, November 10-12, 2000.
Pullan, M. R., Watson, M. F., Kennedy, J. B., Raguenaud, C & Hyam, R. 2000. The Prometheus Taxonomic Model: a practical approach to representing multiple classifications. Taxon 49:55-75.
Roskov, Y. & Bisby, F. 2004 [11 Mar]. Species 2000: an architecture and strategy for creating the Catalogue of the World's Plants. [http://www.sp2000.org/presentations.html].
Scoble, M.J. 2004. (In press). Unitary or unified taxonomy? In Taxonomy for the 21st century Godfray, H. C. J. & Knapp, S. (eds,) Philosophical Transactions of the Royal Society (Biological Sciences).
Wisskirchen, R. & Haeupler, H. (ed.) 1998. Standardliste der Farn- und Blütenpflanzen Deutschlands. Ulmer.
Ytow, N., Morse, D. R. & Roberts, D. M. 2001. Nomencurator: a nomenclatural history model to handle multiple taxonomic views. Biol. J. Linn. Soc. 73:81-98.
Zhong, Y., Jung, S., Pramanik, S. & Beaman, J. H. 1996: Data model and comparison and query methods for interacting classifications in a taxonomic database. Taxon 45: 223-241.
| BGBM Überblick / Overview | Abt.
Biodiversitätsinformatik / Dept. of Biodiversity Informatics |
BGBM Index |
© 2005 Freie Universität
Berlin, Botanischer Garten und Botanisches Museum Berlin-Dahlem,
Seitenverantwortlicher / Page editor: W.
G. Berendsohn.
BGBM Impressum / Imprint
Disclaimer / Haftungsausschluss